Project
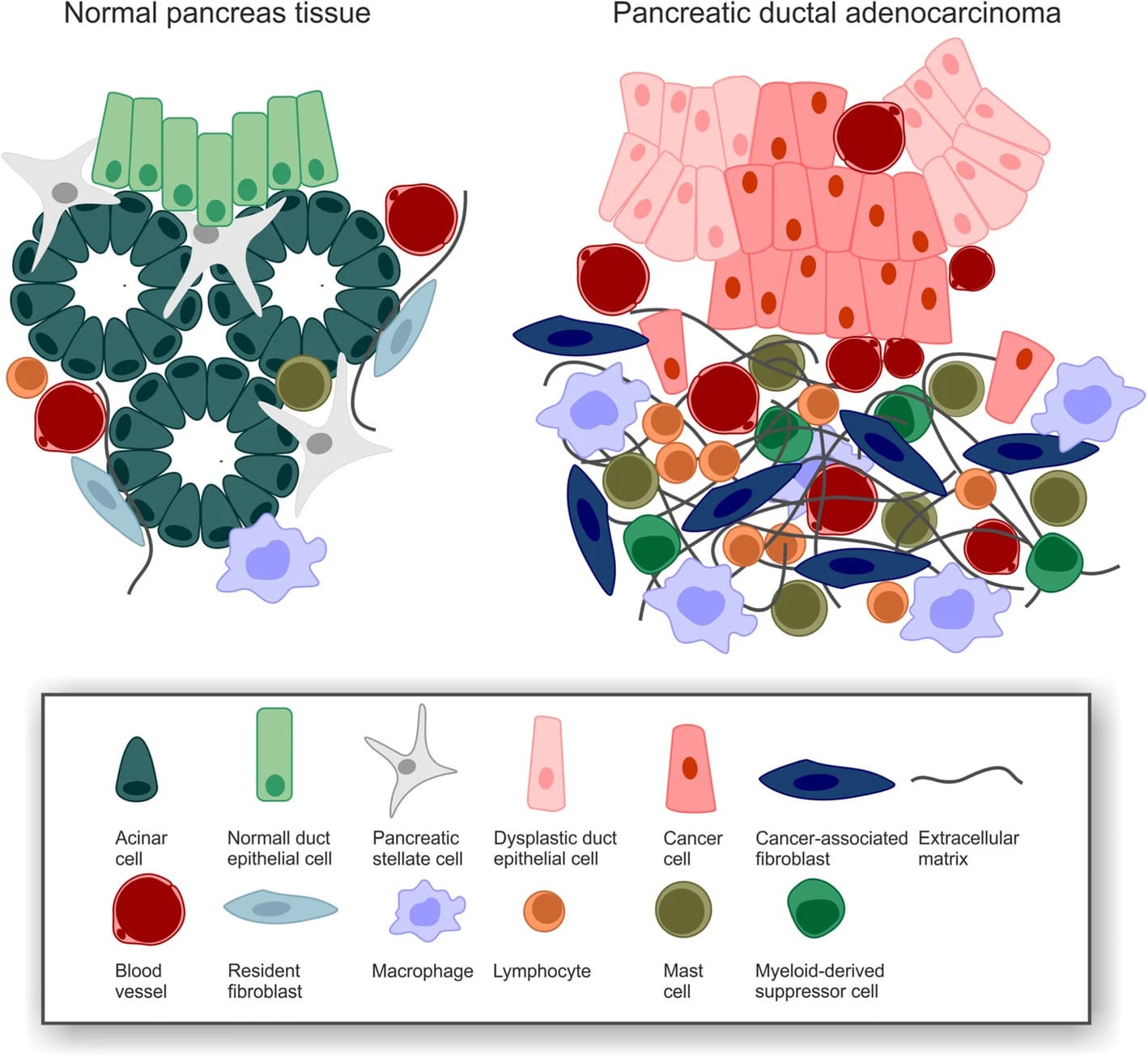
PDAC desmoplasia https://doi.org/10.1186/s13014-019-1345-6
Aim 1:
Comprehensive, integrated genomic analysis of pancreatic cancers with matched normal tissue samples from Indian cohorts using a combination of whole-genome exome and RNA sequencing.
Pancreatic cancer [Pancreatic ductal adenocarcinoma (PDAC)] is an important public health problem and is the fourth leading cause of cancer death worldwide with little improvement in outcomes despite decades of research. It is among the cancers with very poor survival rates. Due to the lack of non-invasive sensitive genomic biomarkers, result in diagnosis often after the cancer has advanced locally to the point of being non-resectable or metastasized to distant sites. At present, there are no specific successful non-invasive detection methods for PDAC. We propose to carry out the following: 1. Comprehensive, integrated genomic analysis of 75 pancreatic cancers with matched normal tissue samples from Indian cohorts using a combination of NGS whole-genome exome and transcriptome sequencing. 2. Determine the mutational mechanisms and candidate genomic events important in pancreatic carcinogenesis. 3. RNA expression analysis to define subtypes and the different transcriptional networks that underpin them.
Aim 2:
To identify specific biomarkers for early detection and novel drug targets to design better therapeutics
Based on the differential expression profile and mutational landscape confirmed using RT-qPCR and Sanger sequencing, the significantly deregulated genes will be selected for developing NGS panel for early detection. Targeted NGS panels will have more clinical advantage over other existing diagnostic methods, due to faster turnaround times and higher sequencing depths resulting in higher analytical sensitivity and specificity. The panel will be designed to interrogate SNVs, indels, CNVs/LOH, and both known and novel fusions associated with major fusion partners. One of the major benefits of using an NGS-based genetic testing strategy is the ability to detect multiple types of aberrations in a single assay. Also, as an alternate, we will develop antibody-based detection assay based on the availability of antibodies. We will test the detection limit of both assays before being validated with a large number of tumor samples.
Expected deliverables of the research
The genomic sequences produced from the proposed work will be critical for developing India’s specific cancer genome database, and this database will be crucial for developing cancer-specific biomarkers for early detection and understanding the treatment response of the patients. Biomarkers identified from the proposed work will be critical for developing Real-time quantitative PCR (RT-qPCR) and sequencing-based early diagnostic kits. Also, identified drug targets will be used for developing novel anti-cancer therapeutics for pancreatic cancer and pediatric leukemia. The preclinical cancer models will be of immense help for drug screening and to understand the in vivo cancer pathogenesis. The successful development of an organoid may replace the animal model for cancer drug screening and genomics. A similar approach will be extended to other cancers that are prevalent in India, and this will help to initiate a start-up company on cancer therapeutics or diagnostics.
Current status
Collected the pancreatic cancer tissue samples from patients after informed consent.
Isolated DNA from cancer tissue and the matched normal tissue samples for exome analysis.
RNA isolation for transcriptome analysis under progress.
Collaborations
International Collaborations
- Established collaboration with Dr. C. Charbel and Dr. Ygal Haupt group at Peter MacCallum Cancer Center, Australia, on cancer genomics and animal model for understanding cancer pathogenesis. Jointly, we have submitted a collaborative proposal to the recently advertised call on Indo-Australian Biotechnology fund (Indian PIs: Dr. S, Mahalingam and Dr. Karthik Raman/Australian PIs: Dr. C. Charbel and Dr. Y. Haupt). Also, initiated the discussion on the exchange of students between the two institutes. We may also attract YIF from Australia.
- Already signed an MOU with Omics Data Automation, USA, and collaboratively working on developing a cancer genome database for the last one year and we have initiated the development of India’s first Breast cancer genome database with 340 breast exome and transcriptome.
- Established collaboration with Dr. K. Rajalingam’s group at University of Mainz, Germany, on cancer pathogenesis on novel drug target identification and already we have published two research articles in high impact international peer-reviewed journals (Journal of Biological Chemistry, 2018a, b).
International education programs, conferences/seminar/webinar links
- This CoE proposal can integrate well with existing and upcoming master’s programmes in computational biology and data science. The novel datasets generated through this study will also have a lot of pedagogical value. Courses related to cancer genomics, machine learning applications for biology can be offered, which will be attractive electives in interdisciplinary programmes already running at IIT Madras, as well as proposed International Master’s programmes.
- To create awareness on cancer genomics and therapeutics to faculties and students from Indian research institutes, universities, and medical institutions, we have already established “CANCERCON” international conference series and successfully conducted CANCERCON 2010, 2014, and 2018 in collaboration with Cancer Research and Relief Trust, Chennai. More than 70 international and national faculties and more than 450 students from all over the world participated. Representatives from many pharma companies have also participated. To the best of our knowledge, this was the only conference in India for which journals like Nature, Science, EMBO, Cell, and European Biochemical Society, sponsored and participated.
- Since there is a lot of demand for cancer genome sequence data analysis experts in India, it is the need of the hour to establish conference and training workshops in this area, the proposed CoE has the potential to create international conference/ workshop series on cancer omics like Gordon/EMBO international conferences.
- We are in discussion with The Cancer Genome Atlas (TCGA), USA, to jointly organize the conferences (every year) on cancer genomics and data analysis at IIT Madras, Chennai.
Industrial collaborations
- Signed an MoU with Omics Data Automation, USA, and collaboratively working on developing a cancer genome database for the last one year. We have completed India’s first Breast cancer genome database with 340 breast cancer whole-genome exomes and transcriptomes.
- Signed an MoU with Indivumed, Germany, and collaboratively working on cancer tissue sample collection for genomics and developing a workflow for automating cancer pathology (tissue immune-histochemistry) and integration of patient electronic health record to genomics to understand the treatment response.
- Bristol Myers Squibb pharma has already approached our group to work collaboratively on the identification of novel drug targets and validation for pediatric leukemia.
- Initiated discussion with Janssen Pharmaceutical company of Johnson and Johnson on developing a diagnostic tool for medical residual disease (MRD) for assessing the drug treatment response for pediatric cancers.
- The proposed CoE will give more visibility of our group on cancer genomics both nationally and internationally and attract many consultancy and collaborations with industries on the development and validation of cancer biomarkers for early detection and to understand the treatment responses, as well as the identification of drug targets for the development of novel therapeutics. Importantly, the cancer organoid model developed from this CoE is a unique and ideal model system for screening drugs for treatments. This model system will potentially help to select the best drugs without trial and error towards personalized cancer treatment. We will be in a unique position compared to all other similar research groups/institutes in India due to the unique cancer genome datasets and the availability of cancer tissue samples for validation. The cancer organoid model will attract many pharma industries for collaboration.
- We have already signed an MOU with Indian Academy of Pediatrics on developing biomarkers for early detection and for designing treatment strategies for better management.
Societal impact
India is on the verge of a cancer epidemic. The sequence data from the proposed CoE will help to develop an India-specific cancer genome database, which is critical to identify and develop biomarkers for early detection and understand the drug response. Also, it will help to identify drug targets for novel therapeutics. Since there is no Indian population-specific cancer genome data available today and all studies are based on what is available from the western population, the patient survival rate is very low compared to western societies due to genomic heterogeneity. The proposed CoE is the first step towards developing an India-specific database.
Since the cancer incidence rate is very high, most pharma companies are on the lookout for Indian-specific cancer genome data to establish biomarkers for specific therapeutics. Results from the proposed CoE will have a great impact on cancer research and drug development in India. Also, it will help to represent India in the International cancer genome consortium for pediatric and pancreatic cancer genome sequences. Notably, the novel datasets, which are India-specific, will be a huge contribution to both the society, the clinical and the research .
Sustenance statement
After the completion of the establishment in different phases, the centre will be fully functional to offer its professional and technical services to the scientific community by catering the applications related to their research on varieties of areas that are mainly focusing on identifying cancer-specific biomarkers, drug discovery, and development from the laboratory to industry level.
The centre can generate the resources through professional training programs, professional technical services, collaborative research programs, public-private-partnership, extension and consultancy activities for the industries, and providing incubation facilities for the BioPharma sector for their optimisation and validation studies.
In this way, the centre can generate money by optimally utilizing the infrastructure. Depending on the Centre’s expertise, the principal companies can use the infrastructure and technical expertise of the personnel to train the existing and prospective customer base and their workforce.
In addition, the centre can facilitate summer/winter schools, training programs, and workshops in related fields. Academic institutions, universities, medical institutions, engineering and technical institutions, biotechnology R&D organizations, and BioPharma industries can optimally utilize this facility. Attempts will be made to generate an intellectual property that could be transferred to industry for commercialization.
Technical/ Scientific Progress
New work done in the project
We have collected cancer with matched adjacent normal tissue samples and blood from 75 pancreatic cancer patients. Isolated tissue DNA and germ line DNA from tissue samples and PBMCs and performed whole exome sequencing analysis. Preliminary analysis suggest occurrence of unique genomic variations in Indian populations. Detailed report is attached
Infrastructure developments
Output
Preliminary data was presented in the webinars
Whole exome Sequencing Analysis of Pancreatic cancers of Indian origin
The QC passed pancreatic cancer and matched adjacent normal tissue DNA and germline DNA library pools from 75 patients were sequenced using Illumina Novaseq 6000 as per the manufacturer’s instructions. The sequence data was processed using fastp to remove low quality bases and adapter sequences. The processed reads were mapped to Human reference genome build hg19 using BWA MEM algorithm. The alignments were sorted and converted to bam files using samtools. Duplicate reads were marked and removed using picard tools. Germline variants were called using Strelka2 variant caller. Somatic variants were called using VarScan2 by using tumor and normal sample pairs. Minimum read counts and p-value cut off were set 10 and 0.01 respectively. Variants were annotated using Ensembl Variant Effect Predictor (VEP). Driver genes were identified by analyzing the somatic variants with MutSigCV. On average 29.7 million paired reads (10GB) data were generated per sample and 99% of the reads were mapped to the reference genome.
Germline Variants:
Germline variant calling of the normal tissue and blood samples resulted in total 121715 variants. SNVs were 118666, indels were 3049, multiallelic variants were 2525 out of which 2263 were multiallelic SNVs. Germline variants were summarized in figure 2.
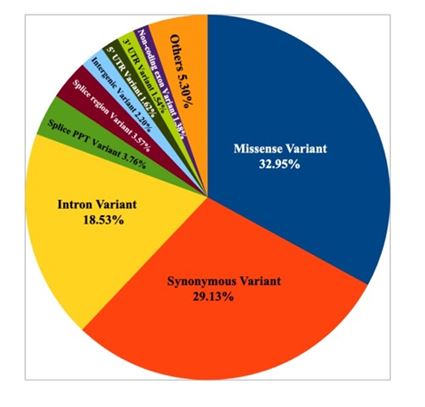
2. Summary of consequences of germline variants.
After filtering the low priority variants 46,544 variants were retained. These variants were further filtered based on genomAD and dbSNP allele frequencies. 14980 variants with allele frequency < 0.01 were retained. In addition, these variants, few variants that were reported to be associated with any type of cancer were retained.
Somatic Variants:
Number of somatic variants for per sample range from 56 to 507. Majority of the somatic variants were missense followed by synonymous and intron variants. The summary of functional somatic variants is provided in figure 3.
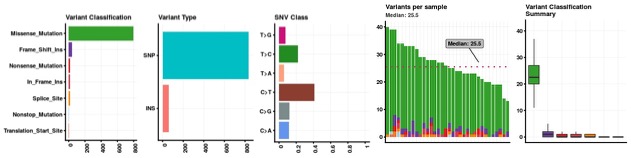
Figure 3. Somatic variant functional classification distributionFigure
Majority of the samples (>93%) have somatic variants in top 10 genes (Figure 4). Mutations in MUC4, DUX4L4 and PRSS3 were present in more than one sample.
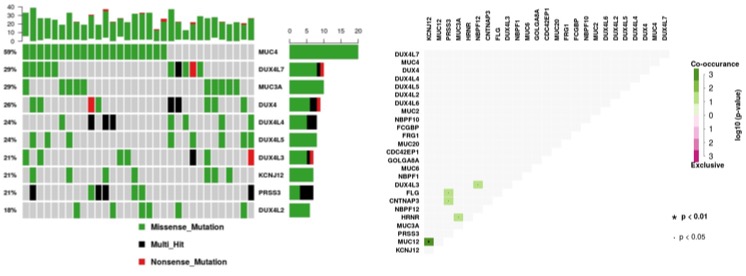
Figure 4. Frequency somatic variants in top 10 genes and somatic co-occurance in pancreatic cancer samples of Indian origin
The change point analysis revealed 5 regions in the genome with relative high number of variants (Table1; Figure 5).
| Table 1. Change points identified in the genome. | |||||
|---|---|---|---|---|---|
| Chromosome | Start | End | nMuts Avg inter-mutation distance | Size | |
| 1 | 12887332 | 13635119 | 28 | 27695.81 | 747787 |
| 1 | 152186422 | 154285522 | 22 | 99957.14 | 2099100 |
| 1 | 168511349 | 247162611 | 13 | 6554271.83 | 78651262 |
| 8 | 16859261 | 16859304 | 10 | 4.78 | 43 |
| 10 | 47087077 | 126678147 | 8 | 11370152.86 | 79591070 |
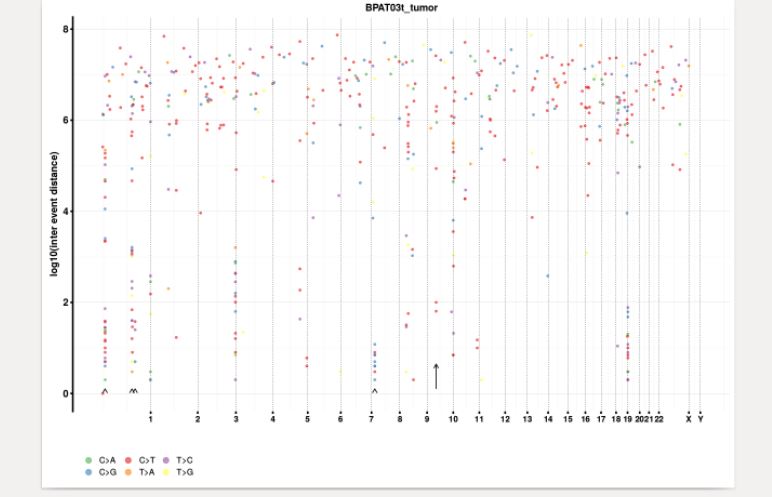
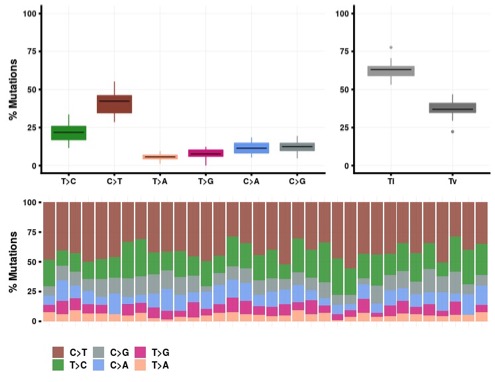
Figure 5. Rainbow plot showing somatic variant distribution and change points in the genome.
Mobility
Visits planned for PI, co-PIs, international collaborators and students (both inbound and outbound)
Relationship
Industrial Engagement
Exploring to develop genomic diagnostics markers and drug screening assays in collaboration with industries for early detection of pancreatic cancers in India.
University Engagement
Establishing collaboration with St.Jude Children’s Hospital, USA for the development of preclinical cancer model for drug screening and understand cancer pathogenesis.
Updates
Relevant Updates
Established the tissue storage, imaging and Next Generation Sequencing facility.
Facilities available at Centre for Cancer Genomics and Molecular Therapeutics
Class 100000 Cancer Tissue Storage facility

Sequencing Facility

Primary Cell Culture Facility

Advanced Imaging Facility





